The research on the fate determination of pluripotent stem cells regulated by single m6A modification was published in Advanced Science by Prof. Nan Cao from Zhongshan School of Medicine and Prof. Guanzheng Luo from the School of Life Sciences
Source: Zhongshan School of Medicine
Edited by: Tan Rongyu, Wang Dongmei
N
6-methyladenosine (m
6A), as one of the most abundant modifications on messenger RNAs (mRNAs), is involved in nearly all the post-transcriptional processes, including RNA splicing, processing, transport, as well as RNA stability and translation efficiency. Previous studies have shown that genetic ablation of the m6A methyltransferase complex METTL3 induces global reduction of m
6A abundance in embryonic stem cells (ESCs) and results in blocked differentiation, suggesting a crucial role of m
6A methylation in regulating early cell fate specification during embryogenesis. Tools for the precise regulation of individual m
6A modifications are lacking in ESCs. Are the deficiencies in cell differentiation arising from a single RNA methylation event or an ensemble of m
6A modifications of multiple sites that function as a synergistic unit? Are modification changes at a single m
6A site sufficient to determine cell fate? These scientific questions remain unanswered.
Prof. Nan Cao's research group from Zhongshan School of Medicine of Sun Yat-sen University, cooperating with Prof. Guanzheng Luo's research team from School of Life Sciences of Sun Yat-sen University, developed a targeted RNA m
6A erasure (TRME) system to achieve site-specific demethylation of RNAs in human ESCs (hESCs) and further demonstrate temporal erasure of a single m
6A site on
SOX2 mRNA could promote ectodermal but inhibits endodermal and mesodermal specification of hESCs.
In this study, the research teams developed a new method for the precise regulation of specific m
6A modification in ESCs, thus laying a foundation for RNA epigenetics studies in ESCs, and providing a new platform for the analysis of stem cell differentiation mechanism, demonstrating that demethylation at a single m
6A site is sufficient to influence the fate of stem cells.
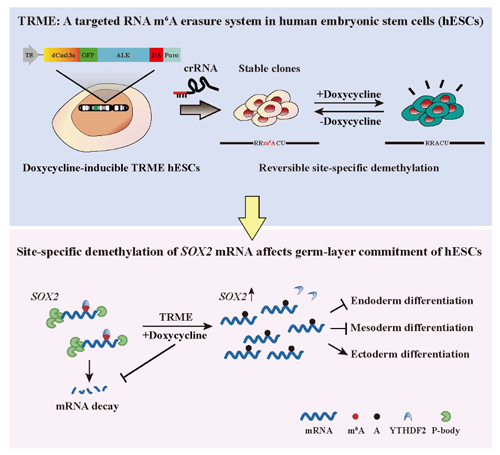
The establishment of the TRME tool and its application in the study of cell fate transition in hESCs
This work entitled "Targeted RNA N
6-methyladenosine Demethylation Controls Cell Fate Transition in Human Pluripotent Stem Cells" was published in the international TOP publication
Advanced Science. Xuena Chen and Qingquan Zhao, PhD students of Zhongshan School of Medicine, and Yuli Zhao, PhD student of School of Life Sciences, are the co-first authors. Prof. Nan Cao and associate researcher Jia Wang of Zhongshan School of Medicine and Prof. Guanzheng Luo of School of Life Sciences are the co-corresponding authors.
Link to the paper:
https://onlinelibrary.wiley.com/doi/10.1002/advs.202003902
