Prof. Wen-Jun Li's group at School of Life Sciences reveal the functional diversity of Chloroflexota from hot springs
Source: School of Life Sciences
Edited by: Zheng Longfei, Wang Dongmei
Chloroflexota, well known for carbon fixation with its unique 3-hydroxypropionate (3-HP) bicycle, is a metabolically diverse and deeply branched bacterial phylum, which play an important role in the biogeochemical cycle on the earth. Although they were widely distributed and abundance in the environment, a large number of Chloroflexota groups still remained uncultured based on molecular ecology studies. Combined with the difficulty in obtaining pure culture, which limited our understanding of their function diversity, it is of great importance for us to gaining insights into this clade by means of culture-independent methods.
Sampling the hot springs from Gujarat and Maharashtra in India, Prof. Wen-Jun Li's group revealed the presence of rod or coccid-rods cells, and the morphologies look similar to those of Chloroflexota members through fluorescence in situ hybridization (FISH) with specific probes (Figure 1a). Seventeen high-quality Chloroflexota metagenome-assembled genomes (MAGs) have been reconstructed using genome-resolved metagenomics, which they might represent novel genus or species based on phylogenomics analysis (Figure 1b). Functional profiling showed that one MAG encoding for bacteriochlorophyll a and Type 2 reaction center (Figure 2), which might be have been transferred from Proteobacteria/Chlorobiota, correlating with earlier studies that indicate phototrophy in the phylum Chloroflexota driven by horizontal gene transfer (HGT). Besides 3-HP bicycle, Chloroflexota might be able to fix carbon via Calvin-Benson-Bassham (CBB) cycle and Wood-Ljungdahl (WL) pathway. The key genes of CBB cycle and WL pathway were detected in some MAGs, but we didn’t notice any HGT related to these pathways. Other key metabolic pathways involved in the MAGs in this study included assimilatory sulfate reduction (sat, cysC and sir), dissimilatory sulfate reduction (dsrAB), dissimilatory nitrate reduction (narGH and nrfAH), urea degradation (ureABC), carbon monoxide oxidation (coxSML and cooSF) and et al. (Figure 2), which greatly expanded the understanding of the functional diversity of Chloroflexota from hot springs. Besides, some members might be motile, indicated by the presence of genes for flagella and chemotaxis. The detection of genes related to heat shock proteins and chaperones in most MAGs suggested that they might be able to maintain their protein machinery stable and efficient in thermal habitats.
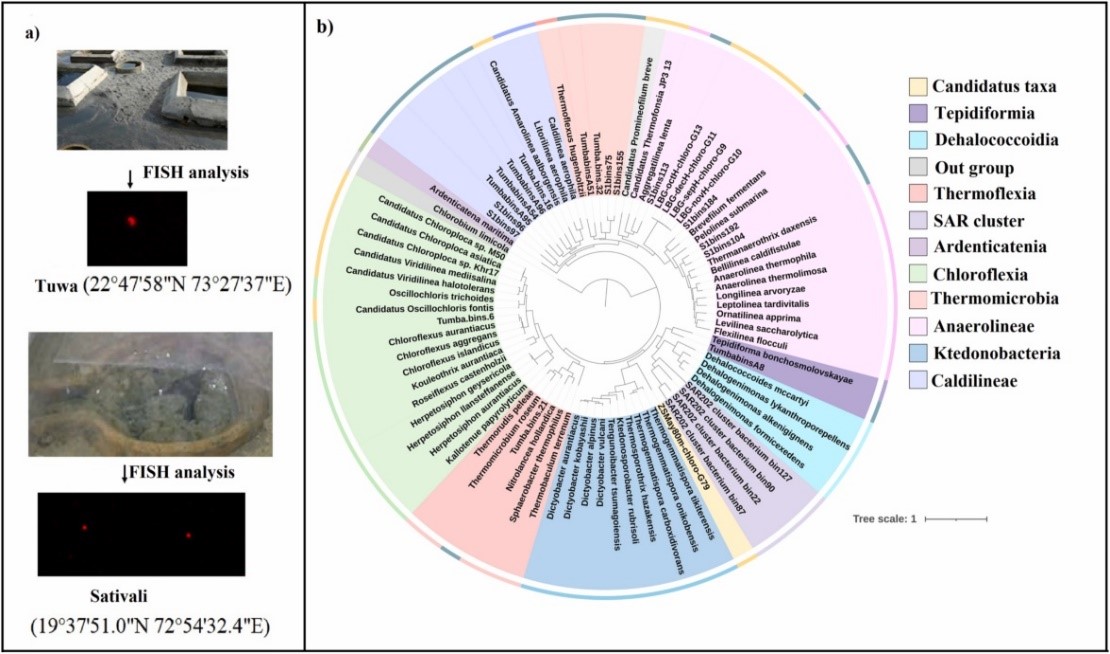
Figure 1 a) geographic location of the hot springs where the samples were collected and FISH analysis showing the precence of Chloroflexota. b), phylogenomics tree reconstruction based on 16 marker genes showing the position of MAGs.
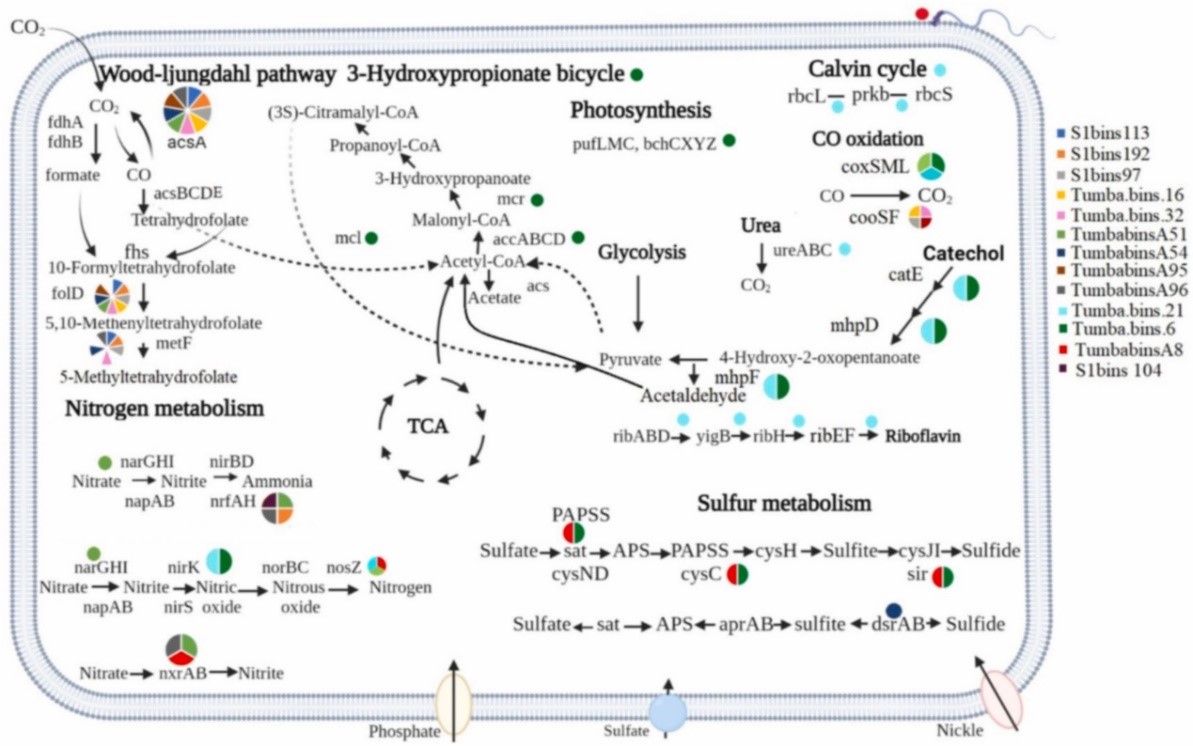
Figure 2 Overview of some key metabolic pathways in Chloroflexota MAGs.
Recently, Prof. Wen-Jun Li's group at the School of Life Sciences, Sun Yat-sen University has published an article titled “Metagenomic analysis further extends the role of Chloroflexi in fundamental biogeochemical cycles” in Environmental Research. Three postdocs from School of Life Sciences in Sun Yat-sen University, Manik Prabhu Narsing Rao, Zhen-Hao Luo and Zhou-Yan Dong are the co-first authors. Professors Wen-Jun Li (from School of Life Sciences in Sun Yat-sen University) and Guo-Xin Nie (from College of Fisheries in Henan Normal University) are the co-corresponding authors. Sun Yat-sen University is the first communication affiliation. This work was financially supported by Key-Area Research and Development Program of Guangdong Province (2018B020206001) and National Natural Science Foundation of China (No, 91951205).
Link to the article: https://doi.org/10.1016/j.envres.2022.112888
