The rediscovery of N6-deoxyadenosine (6mA) in eukaryotic genomes has sparked interest in its potential as a new epigenetic marker. In recent years, 6mA has been identified in various eukaryotes, including plants, vertebrates, and mammals. However, limitations in current methods, particularly the inability to accurately identify the location of 6mA, have hindered further research into its mechanisms and sparked debates about its existence and function in eukaryotes. It is therefore essential to develop a sensitive and reliable method to confirm the existence of 6mA and understand its role in biological processes.
Guan-Zheng Luo, a professor at the School of Life Sciences at Sun Yat-sen University, has spent much of his career studying epigenetics. His earlier work established 6mA as a potential new epigenetic marker (Cell, 2015) and provided insights into its potential functions (Nat Rev Mol Cell Bio, 2015; Nat Struc Mol Bio, 2017). Recently, Luo's group developed a high-resolution method called MM-seq (Modification-induced Mismatch Sequencing) for genome-wide 6mA mapping using a novel detection principle. With this method, the group identified the distribution of 6mA in bacteria (E. coli), green algae (C. reinhardtii), and mammalian cells (HEK239T, Huh7, and HeLa cells) and found a very limited number of 6mA sites scattered across the genomes of different human cell types. Additionally, they observed a significant decrease in the number of 6mA sites after knocking out METTL3, an RNA m6A methyltransferase.
In this article, the authors provide evidence that the m6A/6mA antibody can capture 6mA which is located in double-stranded DNA. They suggest that 6mA may disrupt base-stacking and cause DNA to be partially unstable. The authors use the KMnO4 footprinting assay to confirm that 6mA has the ability to form a mismatch-like structure on duplex DNA, possibly through DNA breathing or base-flipping mechanisms. This mismatch-like structure allows the m6A/6mA antibody to access and bind to the 6mA, and the structure can then be identified and cleaved by specific nucleases or chemicals. Based on this principle, the authors have developed the MM-seq method for precise, genome-wide 6mA mapping, overcoming current limitations in 6mA detection.
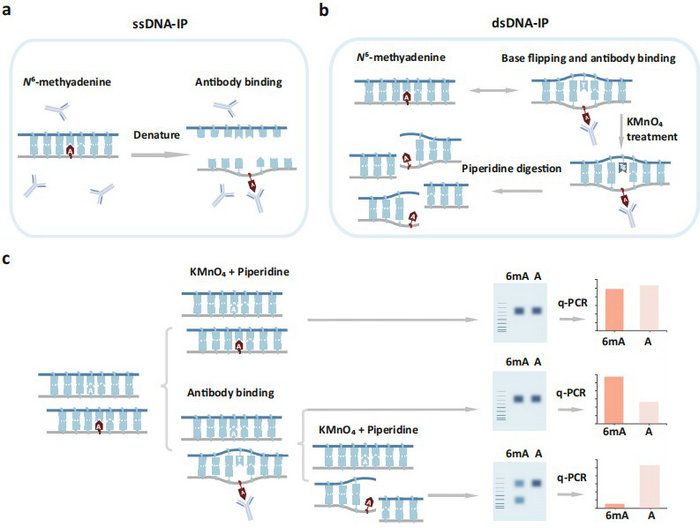
Figure 1. KMnO4 footprinting detects 6mA-induced mismatch-like DNA structure
The authors then used the high resolution and precision of the MM-seq method to study the presence of 6mA in mammalian cells, which have previously been difficult to detect due to their extremely low levels. They found that the 6mA sites identified were very few and scattered randomly across the human genome, with little overlap between different cell types. Despite the low abundance of 6mA, it is possible that it accumulates in specific locations to play a vital role. Therefore, the authors examined whether 6mA tends to cluster in specific regions. Contrary to previous research, they found no enrichment of 6mA in genomic or mitochondrial repetitive elements, indicating that previous IP-based methods may have a high false-positive rate.
In support of the idea that 6mA is formed from the recycling of modified adenosine from RNA degradation, the authors found that the presence of 6mA decreased even further in METTL3-knockout mES cells.
This work has been published in Cell Discovery and entitled as "High-precision mapping reveals rare N6-deoxyadenosine methylation in the mammalian genome". Drs. Li-Qian Chen and Zhang Zhang are co-first authors, with Dr. Luo as corresponding author. Other team members include Hong-Xuan Chen, Jian-Fei Xi, Xue-Hong Liu, Dong-Zhao Ma, Yu-Hao Zhong, Wen Hui Ng, and Tao Chen. The work was also supported by Daniel W. Mak at The University of Hong Kong and Qi Chen and Yao-Qing Chen at Sun Yat-sen University's School of Public Health in Shenzhen. A patent has been filed for the methods used in this research.
Link to the paper: https://www.nature.com/articles/s41421-022-00484-1
