Source: Zhongshan Ophthalmic Center
Written by: Zhongshan Ophthalmic Center
Edited by: Tan Rongyu, Wang Dongmei
Single-cell RNA-sequencing (scRNA-seq) can provide insights into cellular and molecular mechanisms underlying development, regeneration, and ageing. However, there is currently no genome-wide method for accurate, efficient, and quantitative analysis of RNA isoforms in single cells.
Recently, the research team of Professor Yizhi Liu and Dr. Youjin Hu from Zhongshan Ophthalmic Center, Sun Yat-sen University developed scRCAT-seq, a cost-effective and efficient approach to demarcate the boundaries of isoforms based on short-read sequencing. By applying scRCAT-seq to profile the single cells of human retina during development, they revealed the dynamics of TSS and TES during RPC differentiation and cell fate determination. The results have been published in
Nature Communications on Oct. 13th, 2020, entitled "Single-cell RNA cap and tail sequencing (scRCAT-seq) reveals subtype-specific isoforms differing in transcript demarcation".
ScRCAT-seq was developed based on well-established short-read sequencing platforms to explicitly exploit transcription initiation and termination sites for RNA isoforms in single cells. When deployed in conjunction with optimized machine learning models, scRCAT-seq is more accurate, cost-effective, and efficient than existing methods in profiling isoforms with alternative TSS/TES choices (Figure 2). Compared with published approaches, scRCAT-seq can significantly improve the accuracy of TSS and TES peaks by 4-5 times.
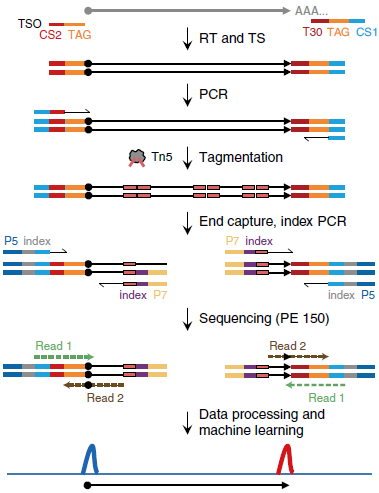
Figure 1. Schematic of the scRCAT-seq.
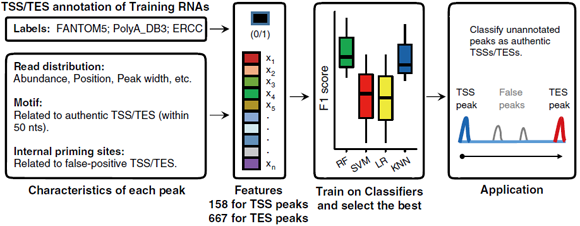
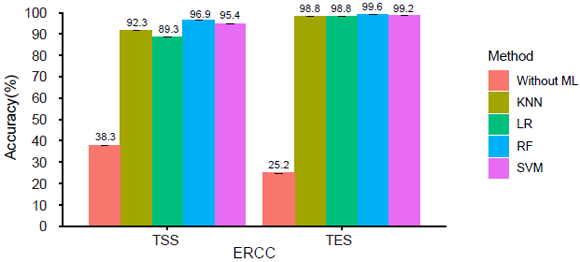
Figure 2. Machine learning model algorithms substantially improved the accuracy of TSS/TES peaks.
Compared with Smart-seq2 and ScISOr-seq, scRCAT-seq exhibited lower cost and higher efficiency. At an equal cost, scRCAT-seq can detect 7 times more transcripts than ScISOr-seq. In addition, scRCAT-seq can detect a large number of genes which ScISOr-seq cannot detect. ScRCAT-seq can detect thousands of novel transcripts and improve the annotation of complex genomes. Coupled with single-cell long-read sequencing or Sanger sequencing, alternative splicing of transcript can further be identified.
Figure 3. The cost-effectiveness and high efficiency of scRCAT-seq.
Dynamics of isoform choices during human photoreceptor cone development
The authors applied scRCAT-seq to hESC-derived retinal organoid (Figure 4). In total, 3,407 and 6,395 single cells were collected for 5’-TSS and 3’-TES analysis respectively, randomly distributed into 6 subtypes. Based on the expression pattern of marker genes, the 6 subtypes were matched to RPC, photoreceptor precursor (PR Precursor), Interneuron precursor (IN precursor), photoreceptor cone, retinal ganglion cells (RGC), and horizontal cell (HC). The differences in TSS/TES choices between different subtypes, and dynamics of isoform switching during cone development from RPC were further assessed. They found that the proximal isoform of CCND1a (truncated CCND1a) is expressed in RPC, and the distal isoform (CCND1a) is expressed in PR precursors and cone. The isoform switch from truncated CCND1a to CCND1a may suggest that CCND1 mediates differential cell-cycle properties between RPC and PR precursors.
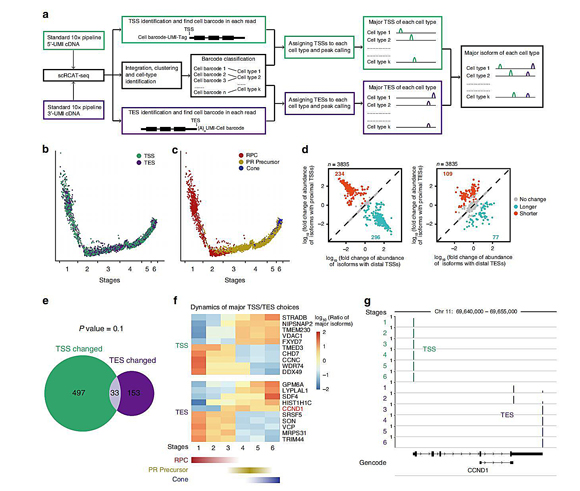
Figure 4. Isoform dynamics during human cone development revealed by scRCAT-seq.
The work was supported by National Key R&D Program of China (2018YFA0108300, 2017YFC1001300); National Natural Science Foundation of China (31700900, 81530028, 81721003); Clinical Innovation Research Program of Guangzhou Regenerative Medicine and Health Guangdong Laboratory (2018GZR0201001); Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program.
Link to the paper:
https://www.nature.com/articles/s41467-020-18976-7



